Last week, all extant phyloreferencers met in sunny Gainesville, Florida for our first face-to-face project meeting. While we’ve met in person before, such as at conferences, this meeting was the first designed specifically to discuss where we are, where we need to be headed, and what therefore our next steps should be. It was also the first meeting to include who will soon be the newest member of the phyloreferencing team: Guanyang Zhang, who will be joining our project as a postdoc at the University of Florida this August!
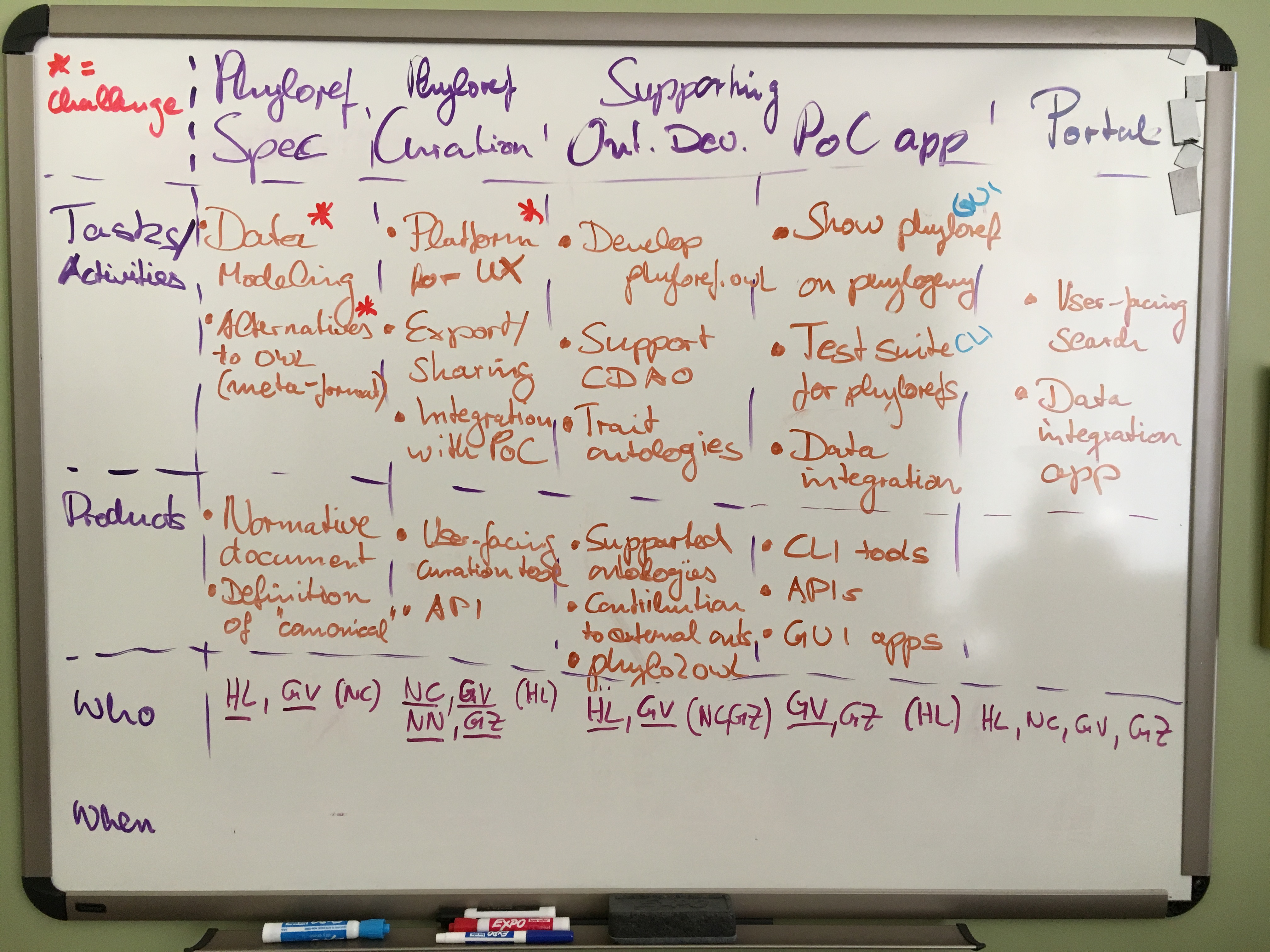
We began by breaking our high-level goals for the next year or two into tasks, activities and products. Finally being in the same room with time to discuss the hard questions of what needs to be done and in which order to accomplish what, we realized that the most important task to group everything else around was developing an effective phyloreference curation workflow. This would start with extracting phylogenetic clade definitions (in natural language) from published articles (“phyloreferences in the wild”), and which would result in a database of testable and visualizable formal phyloreference definitions. Continuously building this database of curated phyloreferences allows us to drive the research and development necessary to determine the best way of modelling phyloreferences in OWL, which in turn will drive developing and refining the concrete phyloreferencing specification. On the tooling side, enabling curators (and so everybody else) to test the phyloreferences they extract will inform the requirements for a proof-of-concept app that allows phyloreferences that don’t behave as expected to be diagnosed and, if necessary, to be corrected easily. Hence, focusing on the curation workflow as the organizing center of our activities will keep us honest (by making sure we meet actual needs!) and will prevent us from going overboard by making sure we don’t get distracted by seemingly cool tools or features whose purpose is poorly defined.
So what should the curation workflow look like? Clearly, it needs to support simultaneous curation from multiple users who have different levels of familiarity with phyloreferences, with OWL and with phylogenies. It needs to produce a diverse set of phyloreferences to test from across a variety of organismal groups. Extracting well-described phylogenetic clade definitions from publications must be easy, as should be extracting and annotating attached phylogenies with the clades described by an author so that well-described test cases can be built from those. A well-described test case for a phyloreference states the specifiers that are to be included, and those that are to be excluded by the clade to which the phyloreference resolves. We can identify these from the tree associated with the phylogenetic clade definition in the original publication. Such a test case can then be evaluated against any tree, including the most encompassing tree of all, the Open Tree of Life. Our next step here is to draw up a detailed plan for the curation workflow that allows us to identify — and subsequently fill — tooling and other gaps. The desired output of this workflow is a set of curated, canonically-represented phyloreferences in an OWL ontology.
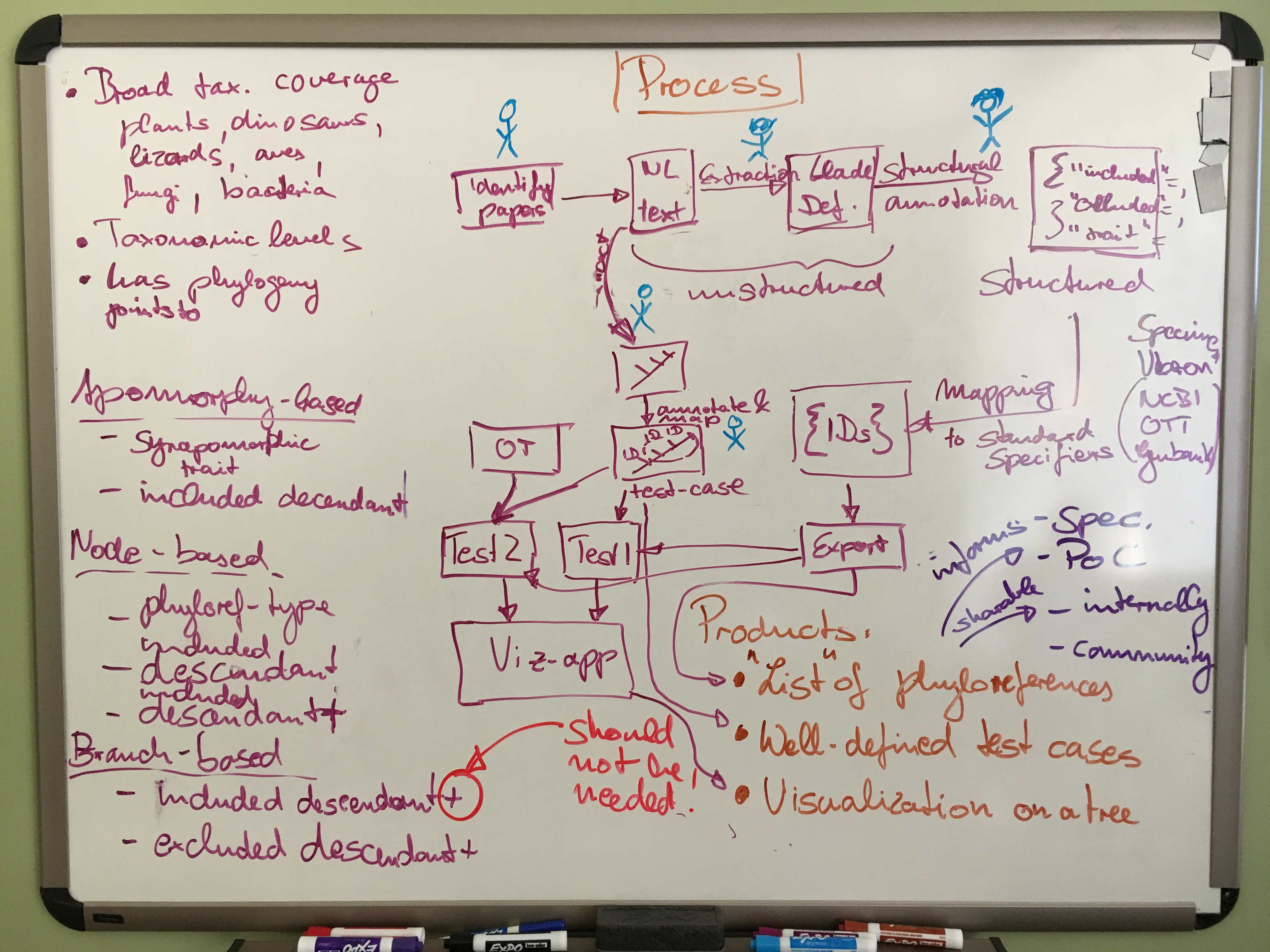
As mentioned above, one of the outcomes we expect from building out and using the curation workflow is to drive research for and development of a phyloreferencing specification, one of the central deliverables of the project. Our emphasis here will be on continuous and rapid iteration cycles, starting with documenting how we code phyloreferences into OWL right now, acknowledging gaps and uncertainties. As we hit the limitations of this initial draft, we’ll iterate towards a more comprehensive and accurate specification. We aim to model phyloreferences in such a way that they’re functionally correct (by matching the clades we’d expect them to match) as well as philosophically sound (by using an accurate ontological representation of phylogenies and clades). We anticipate that this work will involve not only further developing the central phyloreferencing ontology, but also improvements to some of the ancillary ontologies we are or will be reusing, from the (currently orphaned) Comparative Data Analysis Ontology (CDAO) to anatomy and trait-related ontologies. To insulate the curation and annotation of phyloreferences from the expected ongoing changes in the data model and specification, we determined that we will need to keep formal phyloreference generation separate from recording the requisite information in a structured format in our curation workflow. One possibility we might consider for achieving this separation is to create a domain-specific language (DSL), but it’s not clear yet whether that’s necessary. Our next step here is to write up what we might call version 0.1 of our specification, and to share it widely for feedback.
Giving the organizing role to the curation workflow also brought focus to conceptualizing and breaking down what the “Phase I” proof-of-concept application should look like. We decided to follow the Unix philosophy of small, single-purpose tools each of which “do one thing and do it well”, and that can be chained together into a workflow. The most sophisticated tool we need to develop is an application to visualize a phyloreference on top of a phylogeny, along with, when validating a phyloreference versus its test case, what was expected to be included and excluded. Our first step here will be researching various existing phylogenetic visualization tools for their suitability to render such annotations, and the formats best suited for including them. It’s worth noting that authors of publications already contrast alternative phylogenetic clade definitions for groups, and hence eventually there well may be a case for on online visualization app that can render multiple phyloreferences on the same phylogeny, as well as the same phyloreference on multiple phylogenies. The latter will certainly be particularly useful for curation!

To help us better understand how we each think about the factors driving and our project forward or (whether potentially or factually) getting in its way, we concluded our meeting with a light-weight SWOT analysis. Among the important take-aways from this exercise was that because of the novelty of our project, which is one of its biggest strengths but also carries high risks, communicating the value of phyloreferences will need to remain on our minds, as will trying to keep potential future barriers to adoption out of our designs, models, and implementations.
Finally, we discussed ideas and possibilities for independent research projects of the two postdocs on the project. We’ll share these and thoughts about them later as they begin to be developed.
In summary, we’re now in a good place to take some big steps over the next months. We’ll soon have a first specification draft to share and an implementation plan for our curation workflow. Onwards and upwards!
