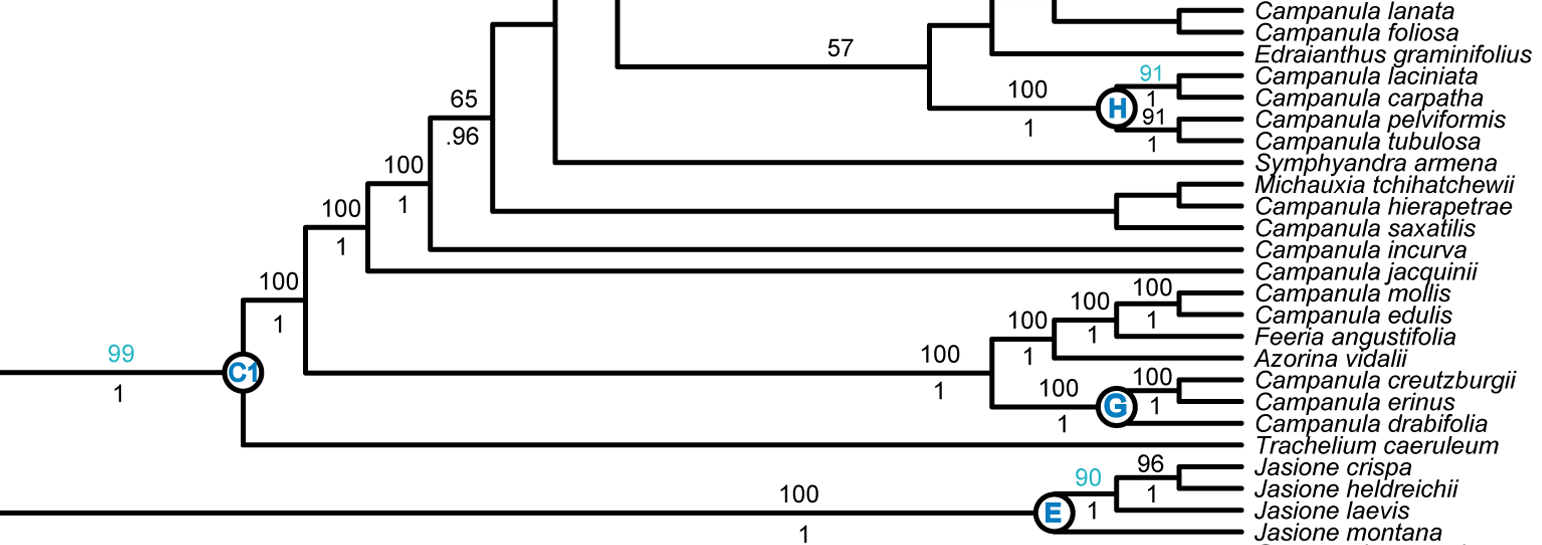
Our current goal for Phyloreferencing is a test suite that will allow us to go from phylogeny to a tested phyloreference with the fewest number of human interactions. To test this, I started with a paper published a few years ago by Andrew A. Crowl and colleagues (including one of our PIs!): Phylogeny of Campanuloideae (Campanulaceae) with Emphasis on the Utility of Nuclear Pentatricopeptide Repeat (PPR) Genes. I wanted to see how many of the twelve named clades in this paper I could represent using Phyloreferencing, and whether those phyloreferences would work across the three phylogenies includes in this paper: plastid tree, pentatricopeptide repeat (PPR) gene family tree, and Plastid + PPR tree. Here’s what that process looked like:
-
The authors submitted their trees as supplementary materials. I started with tree S22, the Plasmid + PPR maximum likelihood tree.
-
Phylo2owl converted the tree to an OWL representation — the generated OWL file passed all our current tests! Trying this extraction made me wonder whether phylo2owl supports files containing identically named leaf nodes; since DendroPy does not, it turns out neither do we. I’ve filed an issue to fix this.
-
I started by trying to create a phyloreference for clade C1 as illustrated in figure 3 of the paper. I planned to create a stem-based definition, which take the form “ancestors of taxon X that are not also ancestors of taxon Y”. One way of doing this is by using a logical construct we call
excludes_lineage_to, which identifies a sibling to some ancestor of the target and that sibling’s descendants, but not ancestors of the target themselves. By using this, we can identify the subclade under C1 (i.e. C1 excluding Trachelium caeruleum) using the following OWL class definition in Manchester syntax:has_Descendant value Campanula_latifolia and excludes_lineage_to value Trachelium_caeruleum -
To find clade C1’s root node, we then need to find the parent of this node. Note that the clade descending from this root node now includes Trachelium caeruleum:
has_Child some (has_Descendant value Campanula_latifolia and excludes_lineage_to value Trachelium_caeruleum) -
For testing purposes, it’s easiest to match the leaf nodes belonging to this clade. So I modified the phyloreference to find every node that has the clade’s root node as an ancestor:
has_Ancestor some (has_Child some (has_Descendant value Campanula_latifolia and excludes_lineage_to value Trachelium_caeruleum))
Tada — my first phyloreference! You can see it in Manchester format on our Github repository as clade_C1. These phyloreferences are easy to test for small clades, where we can enumerate every node we expect to be included in this clade: you can see an example of that in our repository, named clade_C1_expected. Our test suite can now reason over the phyloreference and ensure that the two classes contain identical lists of individuals. Internal nodes are hard to check, but we can ensure that only the correct leaf nodes are included – and they are!
I extended this to two other trees in the paper – tree S20, the Plasmid tree and tree S21, the PPR tree. I immediately ran into problems: some of the first phyloreferences I came up with refer to leaf nodes that are found in one of the trees but not the others. I’ve summarized the differences in the phyloreferences as they currently stand, as well as links to all phyloreferences currently in our test suite. These are the sorts of challenges we will be facing in developing useful phyloreferences, so running into them in this small experiment is a great start!
The next step is to build better phyloreferences – ones that will work across all three of the phylogenies we have currently included, based on the different leaf nodes present in each of those trees. New phyloreferences are on their way!
Image credit: extract of figure 3 from Crowl et al., 2014, available under a CC-BY license.